We are building a team of biologists, engineers
and coders passionate about understanding
how human tissues and organs are made.
Overview
Each fold, layer, and boundary of our organs is the record of a cellular decision: when to grow, where to signal, what to become. Recent advances integrating insights from vertebrate embryonic development with cutting-edge human stem cell technologies have enabled the creation of in vitro cellular systems that recapitulate key steps of complex tissue formation. Our work builds on these foundations to achieve biological tissue design with intent and precision — essential for improving models of human disease and drug discovery.
Meet our team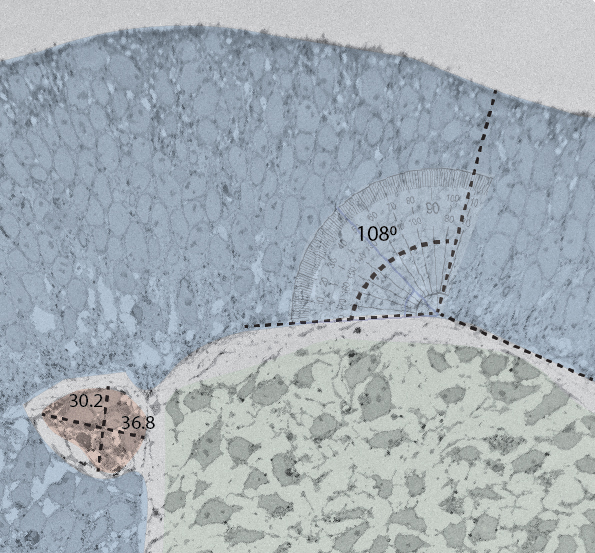
Research projects
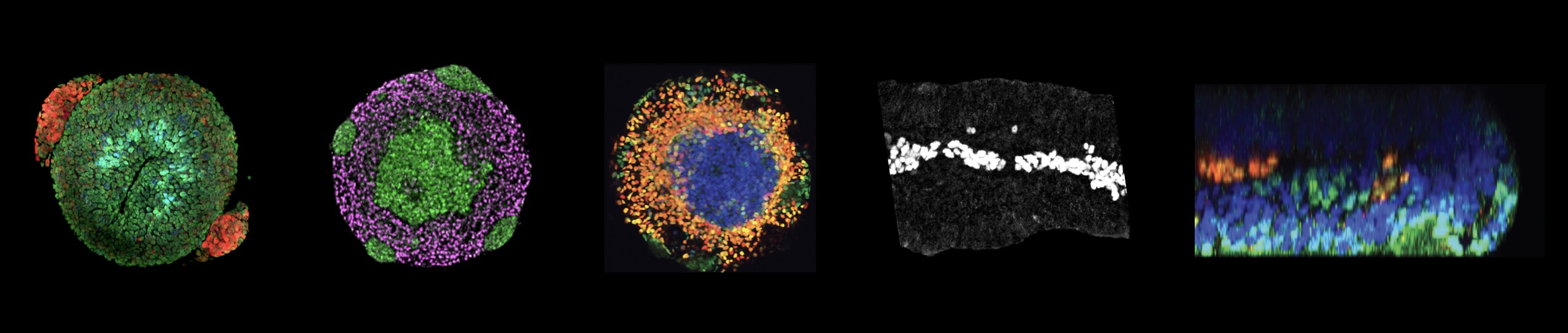
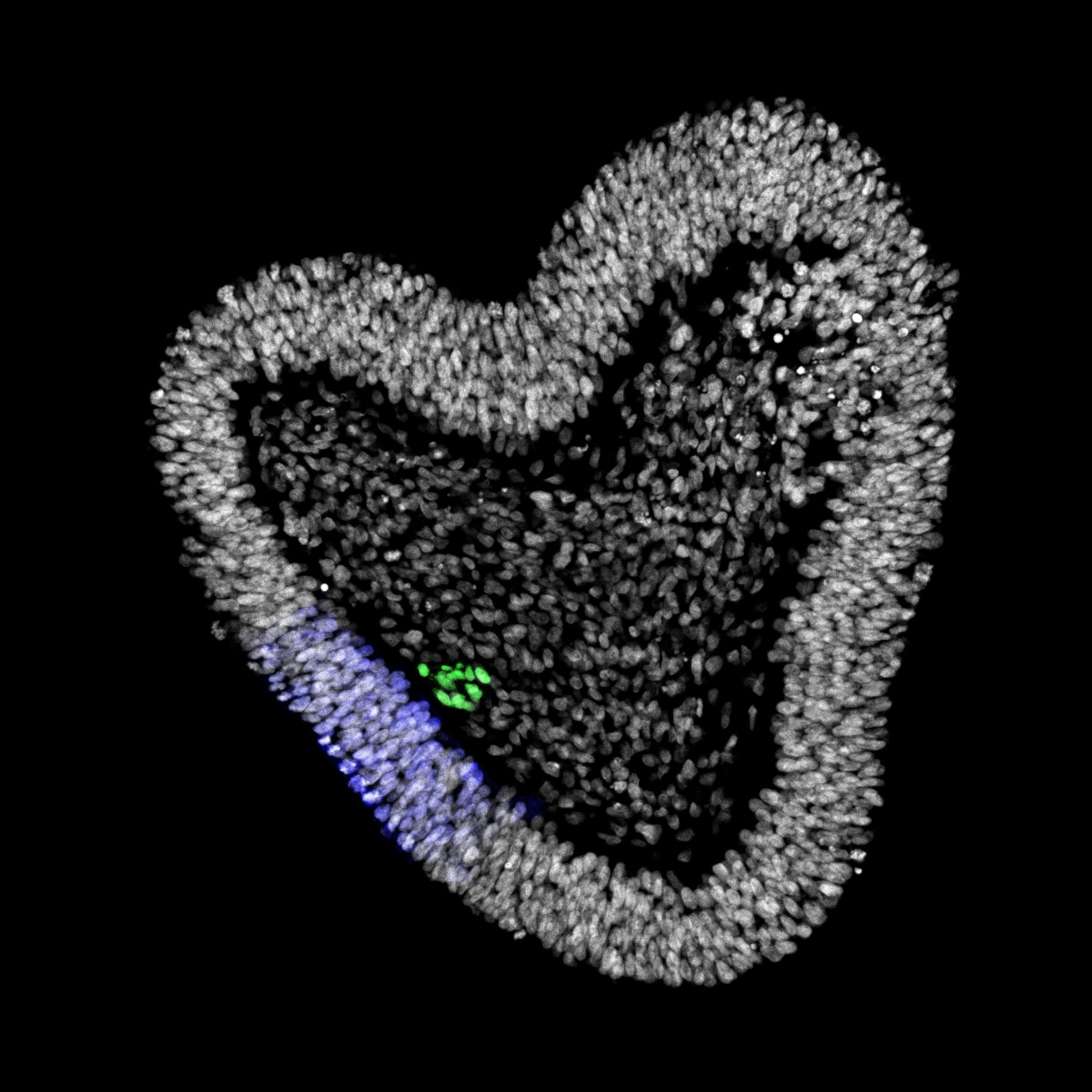
How do cells build organs?
This central question in Developmental Biology touches fundamental principles that govern how a single fertilized egg gives rise to a multicellular organism with diverse, structured tissues and organs. Our research aims to unravel and control the mechanisms governing human cell differentiation, tissue patterning, growth, and maturation.
Mechanism
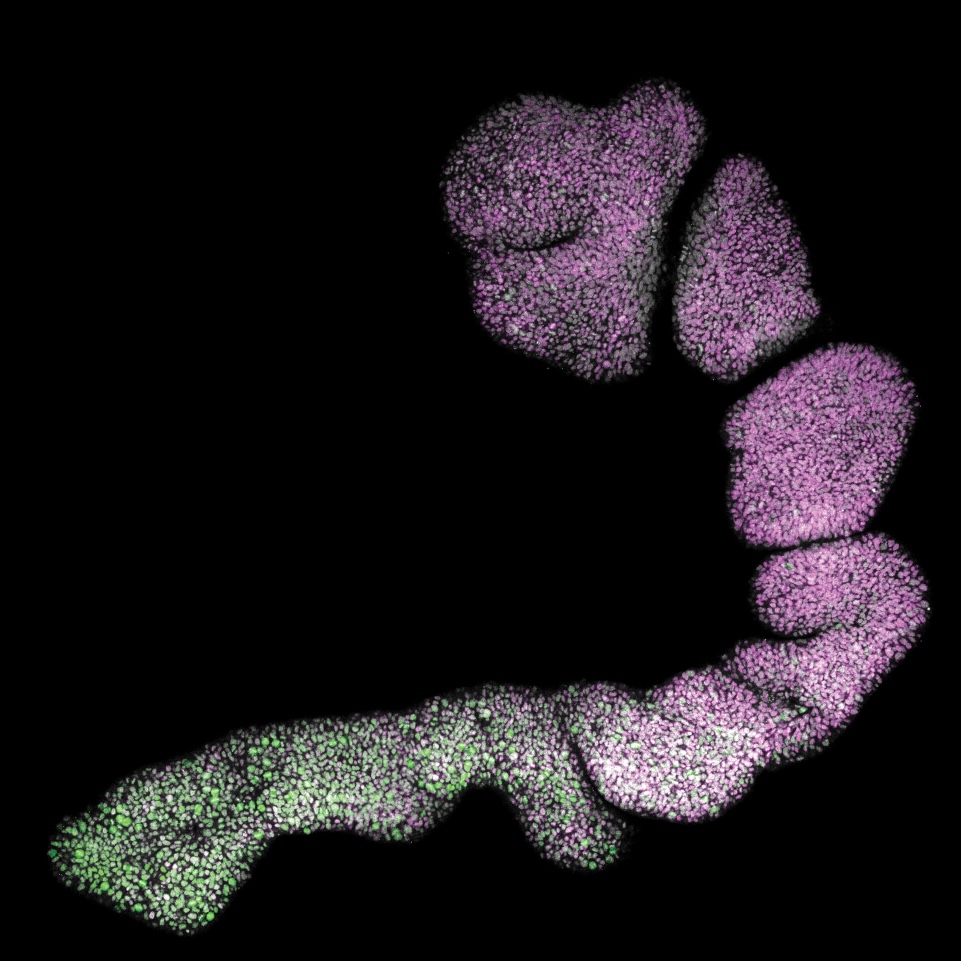
The coordination of timing and intensity of well-conserved signalling pathways, together with cytoskeleton dynamics, underlies many morphological processes that shape our organs. We previously identified the timing of NODAL signalling as crucial for the simultaneous induction of three key embryonic cell populations in the trunk, raising new questions about how cellular decisions emerge in trunk organoids. By combining in silico predictions with experimental validation, we are uncovering the molecular underpinnings of symmetry breaking and the formation of distinct multicellular signalling domains that can pattern and expand at scale into tissues with controlled size and complexity.
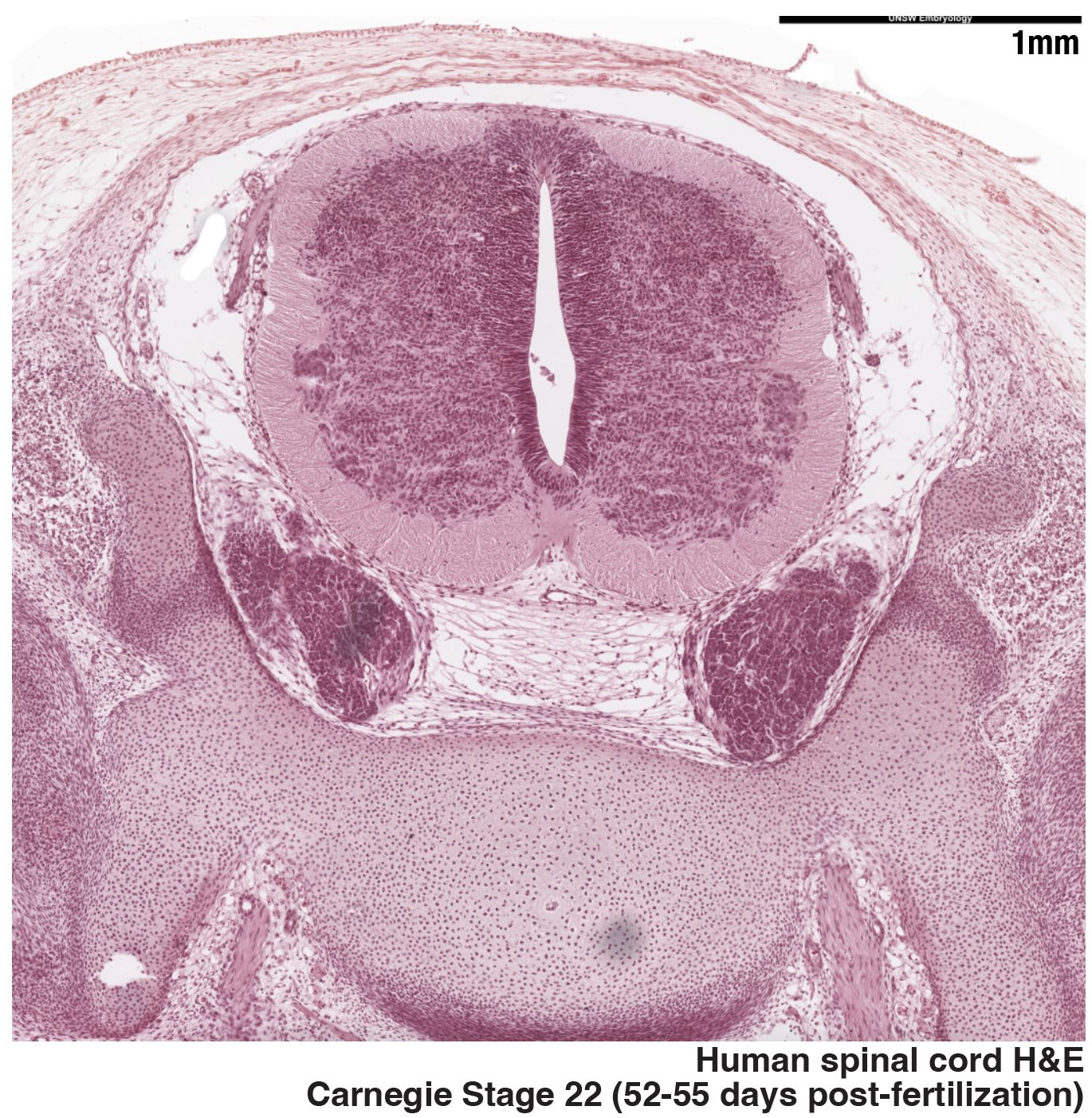
Maturation
The maturation of stem-cell-based embryo models with multiple embryonic germ layers offers a more holistic approach to model human development. Further research will enable more relevant and effective testing of drug targets and disease biomarkers, while also providing a foundation for developing regenerative medicine strategies for diseases such as intervertebral disc degeneration, scoliosis or chordomas. We are characterizing the cellular and molecular changes occurring in the synthetic notochord over time and in different conditions while monitoring the effects on the adjacent neural and mesodermal populations.
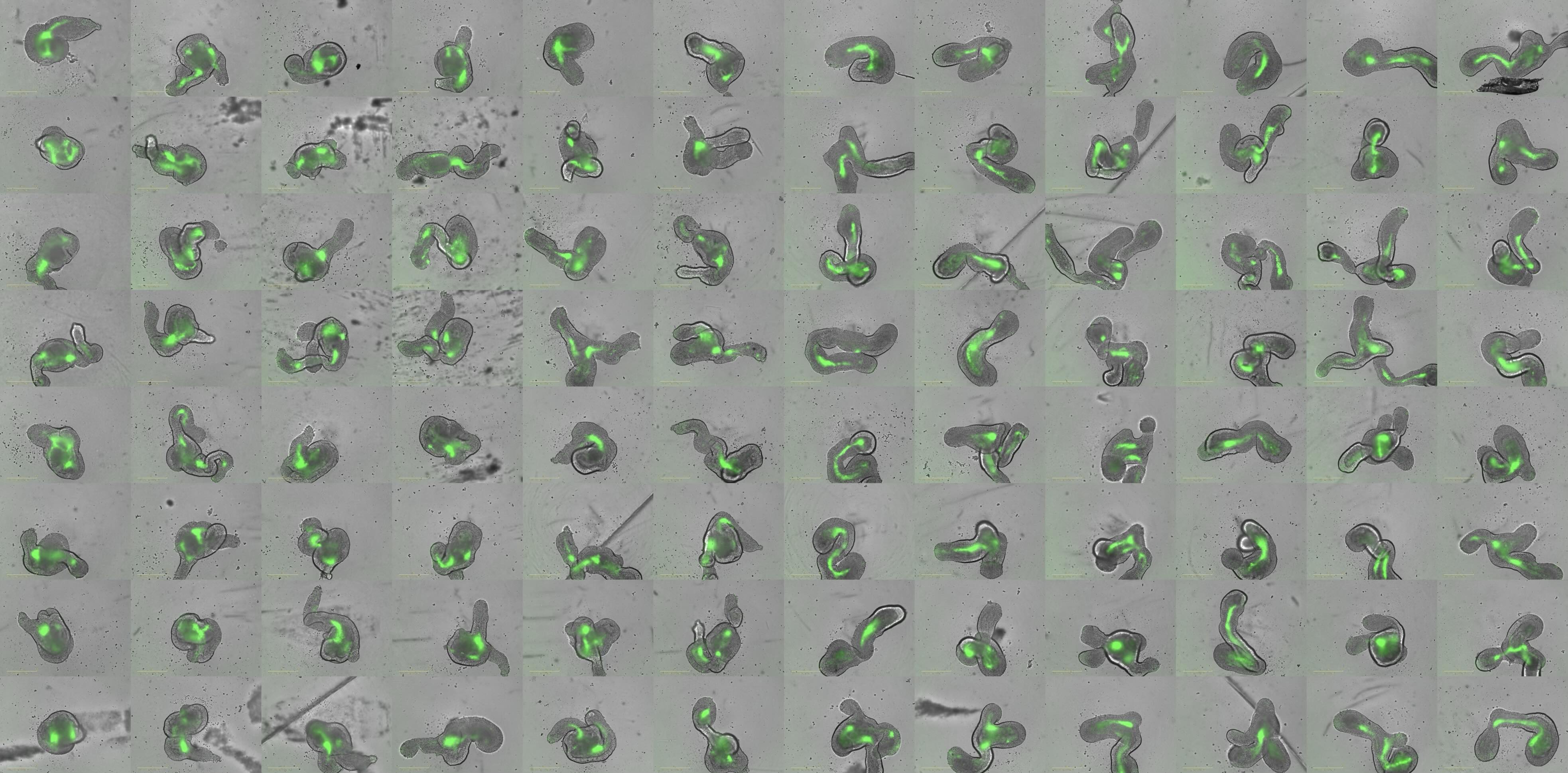
Modification
Understanding the molecular mechanisms behind cellular decisions opens new avenues for controlling organoid morphology and cell fate. By bioengineering specific early tissue architectures, we aim to elucidate how distinct signalling centres can be combined to generate a collection of tunable trunk organoids with defined cell compositions and targeted applications.

Publications and Resources
•
2025
- Tiago Rito, A. R. G. Libby, M. Demuth, M.-C. Domart, J. Cornwall-Scoones and James Briscoe (2025), Timely TGF-β signalling inhibition induces notochord, Nature.
- A.R.G. Libby, Tiago Rito, A. Radley and James Briscoe (2025), An in vivo CRISPR screen in chick embryos reveals a role for MLLTS in specification of neural cells from the caudal epiblast, Development 152 (3): DEV204591.
2021
- M. Yang*, Tiago Rito*, J. Metzger, J. Naftaly, R. Soman, J. Hu, D.F. Albertini, D.H. Barad, Ali H. Brivanlou and Norbert Gleicher (2021), Depletion of aneuploid cells in human embryos and gastruloids, Nature Cell Biology 23: 314–321.
2019
- T. Haremaki*, J. J. Metzger*, Tiago Rito*, M. Z. Ozair, E. Siggia, F. Etoc and Ali H. Brivanlou (2019), Self-organizing human neuruloids allow disease modeling in the developing ectodermal compartment, Nature Biotechnology 37(10): 1198–1208.
2018
- M. Z. Ozair, C. Kirst, B. L. van den Berg, A. Ruzo, Tiago Rito and Ali H. Brivanlou (2018), hPSC Modeling Reveals that Fate Selection of Cortical Deep Projection Neurons Occurs in the Subplate, Cell Stem Cell 23:1 (60–73.e6).
2017
- C. Ferrai, E. Torlai Triglia, J. R. Risner-Janiczek, Tiago Rito, O. J.L. Rackham, I. de Santiago, A. Kukalev, M. Nicodemi, A. Akalin, M. Li, M. A. Ungless and Ana Pombo (2017), RNA polymerase II primes Polycomb-repressed developmental genes throughout terminal neuronal differentiation, Molecular Systems Biology 13: 946.
2015
- J.D. Dias, Tiago Rito*, E. Torlai Triglia*, C. Ferrai, A. Kukalev, M. Chotalia, E. Brookes, H. Kimura and Ana Pombo (2015), Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells, eLife 10.7554/eLife.11215.
- J. Fraser*, C. Ferrai*, A. M. Chiariello*, M. Schueler*, Tiago Rito*, G. Laudanno*, M. Barbieri, B. L. Moore, D. Kraemer, S. Aitken, S. Q. Xie, K. J. Morris, M. Itoh, H. Kawaji, I. Jaeger, Y. Hayashizaki, P. Carninci, A. R.-R. Forrest, the FANTOM Consortium, C. A. Semple, J. Dostie, Ana Pombo, and Mario Nicodemi (2015), Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation, Molecular Systems Biology 11: 852.
2014
- W. Ali, Tiago Rito, G. Reinert, F. Sun and Charlotte M. Deane (2014), Alignment-free protein interaction network comparison, Bioinformatics 30 (17): i430–i437.
2012
- Tiago Rito, C. M. Deane and Gesine Reinert (2012), The importance of age and high degree, in protein-protein interaction networks, Journal of Computational Biology 19 (6): 785–795.
2011
- C. Katz, L. Levy-Beladev, S. Rotem-Bamberger, Tiago Rito, S. G. D. Rüdiger and A. Friedler (2011), Studying protein-protein interactions using peptide arrays, Chem. Soc. Rev. 40(5): 2131–2145.
2010
- Tiago Rito, Z. Wang, C. M. Deane and Gesine Reinert (2010), How threshold behaviour affects the use of subgraphs for network comparison, Bioinformatics 26 (18): i611–i61.6
Our Team
•


Tiago Rito
Assistant Professor
B.Sc. Uni of Coimbra, Portugal – Biochemistry
D.Phil. Uni of Oxford – Statistics
Postdoc: Pombo Lab – Brivanlou Lab – Briscoe Lab
🇵🇹➝🇳🇱➝🇬🇧➝🇩🇪➝🇺🇸➝🇬🇧➝🇭🇰
Tiago has always enjoyed being in the lab, and his postdoctoral years were no exception. For his final postdoc, he joined Dr. James Briscoe’s group in London, where he discovered that it was indeed possible to do everything all at once in the pursuit of a scientific question. He continues to combine classic embryology, human stem cell biology, and computational approaches to study human development. His group focuses on engineering signalling centers in vitro to generate human tissues with defined complexity and morphology, aiming to create reproducible and functional models for research and medicine.


Lab life

Visuals
Connect

Join us!
We are actively looking for passionate and motivated students, postdocs, and research assistants to join our team. If you are curious about how human tissues and organs are formed — and eager to contribute to cutting-edge human stem cell and developmental biology research — we would love to hear from you.
Interested applicants with a strong commitment to science are encouraged to get in touch for more information. Please send an email with the subject line “PhD/Postdoc/RA application” and include:
- Your CV
- A short research statement highlighting your interests and future goals
Prospective graduate students should also consult the university's application roadmap, which includes information on fellowships and scholarships: HKU Graduate School Application Guide.